Deep Dive: Neuron Morphology Analysis#
This comprehensive tutorial explores the morphological analysis capabilities of CRANTpy. We’ll cover:
Mesh representations - Fetching and analyzing neuron meshes
Skeleton representations - Multiple skeletonization methods
L2 graph-based skeletons - High-quality morphology from L2 chunks
Dotprops - Point cloud representations for fast analysis
Morphometric analysis - Cable length, surface area, volume, branching
Soma detection - Automated soma identification
Visualization - 2D and 3D visualization techniques
Population analysis - Comparing morphology across neuron groups
Navis integration - Leveraging navis for advanced analysis
Let’s start by setting up our environment.
# Import CRANTpy and other necessary libraries
import crantpy as cp
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import navis
import cloudvolume as cv
import networkx as nx
# Set up logging
cp.set_logging_level("WARNING")
print("CRANTpy loaded successfully!")
print(f"Default dataset: {cp.CRANT_DEFAULT_DATASET}")
print(f"Navis version: {navis.__version__}")
CRANTpy loaded successfully!
Default dataset: latest
Navis version: 1.10.0
1. Authentication Setup#
First, ensure you are authenticated with the CAVE service.
# Generate and save authentication token (uncomment if first time)
# cp.generate_cave_token(save=True)
# Test connection
try:
client = cp.get_cave_client()
print(f"Successfully connected to datastack: {client.datastack_name}")
print(f"Server: {client.server_address}")
except Exception as e:
print(f"Connection failed: {e}")
print("Please run: cp.generate_cave_token(save=True)")
Successfully connected to datastack: kronauer_ant
Server: https://proofreading.zetta.ai
2. Getting Sample Neurons#
Let’s query some neurons for our morphology analysis.
# Get a sample of olfactory projection neurons
opn_criteria = cp.NeuronCriteria(cell_class='olfactory_projection_neuron')
opn_ids = opn_criteria.get_roots()
print(f"Found {len(opn_ids)} olfactory projection neurons")
# Select a few for detailed analysis
sample_ids = opn_ids[:3]
print(f"\nSample neurons for detailed analysis:")
for i, neuron_id in enumerate(sample_ids, 1):
print(f" {i}. {neuron_id}")
Found 107 olfactory projection neurons
Sample neurons for detailed analysis:
1. 576460752700282748
2. 576460752681552812
3. 576460752722405178
3. Mesh Representations#
Meshes provide the most detailed representation of neuron morphology, capturing the full 3D surface.
Fetching Mesh Neurons#
# Fetch a single mesh neuron
single_mesh = cp.get_mesh_neuron(sample_ids[0])
print(f"Mesh neuron properties:")
print(f" ID: {single_mesh.id}")
print(f" Vertices: {len(single_mesh.vertices):,}")
print(f" Faces: {len(single_mesh.faces):,}")
print(f" Volume: {single_mesh.volume:.2f} cubic nanometers")
Mesh neuron properties:
ID: 576460752700282748
Vertices: 238,432
Faces: 478,609
Volume: 135639679325.33 cubic nanometers
# Fetch multiple mesh neurons
mesh_neurons = cp.get_mesh_neuron(sample_ids, threads=3, progress=True)
print(f"\nFetched {len(mesh_neurons)} mesh neurons")
# Summary of meshes
for n in mesh_neurons:
print(f" {n.id}: {len(n.vertices):,} vertices, {n.volume:.2f} μm³")
Fetched 3 mesh neurons
576460752700282748: 238,432 vertices, 135639679325.33 μm³
576460752681552812: 309,633 vertices, 192266769082.67 μm³
576460752722405178: 248,290 vertices, 154247730149.33 μm³
576460752681552812: 309,633 vertices, 192266769082.67 μm³
576460752722405178: 248,290 vertices, 154247730149.33 μm³
Soma Detection#
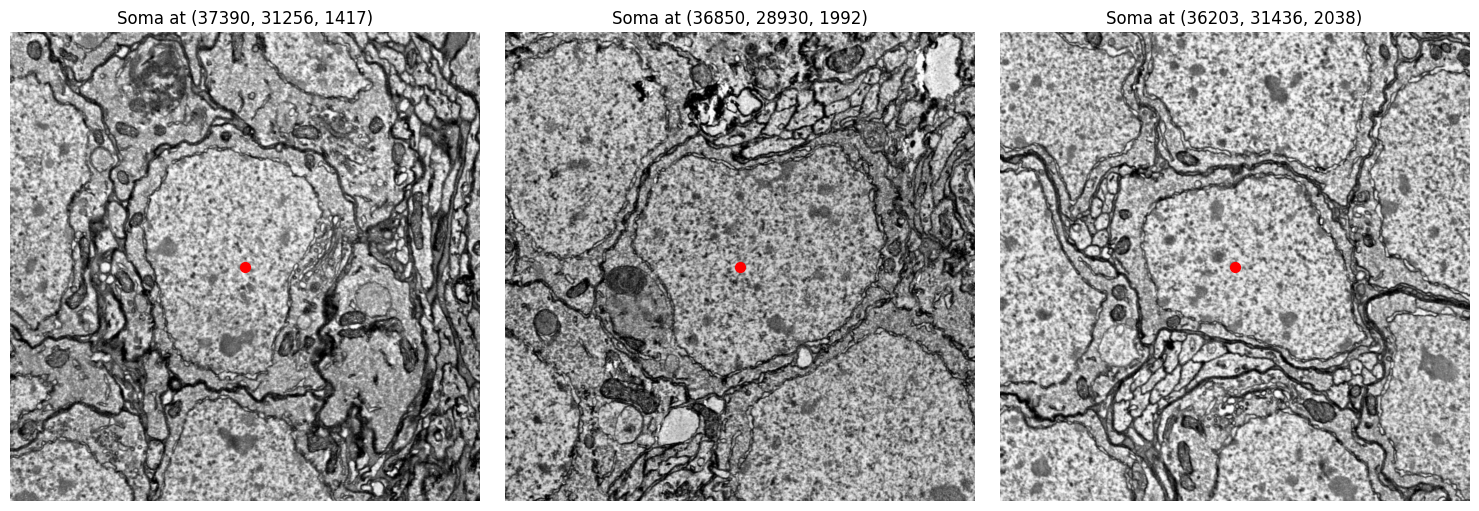
We can use the function detect_soma to retrieve the estimated pixel coordinates of a neuron’s soma. We can then use a helper function to visually inspect this area in the raw EM data to verify it contains a nucleus.
# Detect soma for a single neuron
soma_coords = cp.detect_soma(sample_ids[0])
print(f"Soma detected at coordinates: {soma_coords}")
# Detect soma for multiple neurons
soma_coords_batch = cp.detect_soma(sample_ids, progress=True)
print(f"\nSoma coordinates for {len(sample_ids)} neurons:")
for neuron_id, coords in zip(sample_ids, soma_coords_batch):
print(f" {neuron_id}: {coords}")
Soma detected at coordinates: [37390 31256 1417]
Detecting soma: 0%| | 0/3 [00:00<?, ?it/s]
Detecting soma: 33%|███▎ | 1/3 [00:05<00:10, 5.05s/it]
Detecting soma: 67%|██████▋ | 2/3 [00:11<00:06, 6.03s/it]
Soma coordinates for 3 neurons:
576460752700282748: [37390 31256 1417]
576460752681552812: [36850 28930 1992]
576460752722405178: [36203 31436 2038]
# Plot EM image around the detected somas
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
for ax, coords in zip(axes, soma_coords_batch):
x, y, z = map(int, coords)
img = cp.plot_em_image(x, y, z, size=1000)
ax.scatter(500, 500, color='red', s=50)
ax.imshow(img, cmap='gray')
ax.set_title(f"Soma at ({x}, {y}, {z})")
ax.axis('off')
plt.tight_layout()
plt.show()
Downloading Bundles: 100%|██████████| 11/11 [00:00<00:00, 12.60it/s]
Downloading Bundles: 100%|██████████| 11/11 [00:00<00:00, 12.60it/s]
Decompressing: 100%|██████████| 25/25 [00:00<00:00, 1637.84it/s]
Shard Indices: 0%| | 0/4 [00:00<?, ?it/s]
Shard Indices: 100%|██████████| 4/4 [00:00<00:00, 33.11it/s]
Minishard Indices: 100%|██████████| 4/4 [00:00<00:00, 34.10it/s]
Minishard Indices: 100%|██████████| 4/4 [00:00<00:00, 34.10it/s]
Downloading Bundles: 100%|██████████| 10/10 [00:00<00:00, 16.33it/s]
Downloading Bundles: 100%|██████████| 10/10 [00:00<00:00, 16.33it/s]
Decompressing: 100%|██████████| 20/20 [00:00<00:00, 1424.79it/s]
Downloading Bundles: 100%|██████████| 11/11 [00:00<00:00, 12.45it/s]
Downloading Bundles: 100%|██████████| 11/11 [00:00<00:00, 12.45it/s]
Decompressing: 100%|██████████| 25/25 [00:00<00:00, 1049.49it/s]
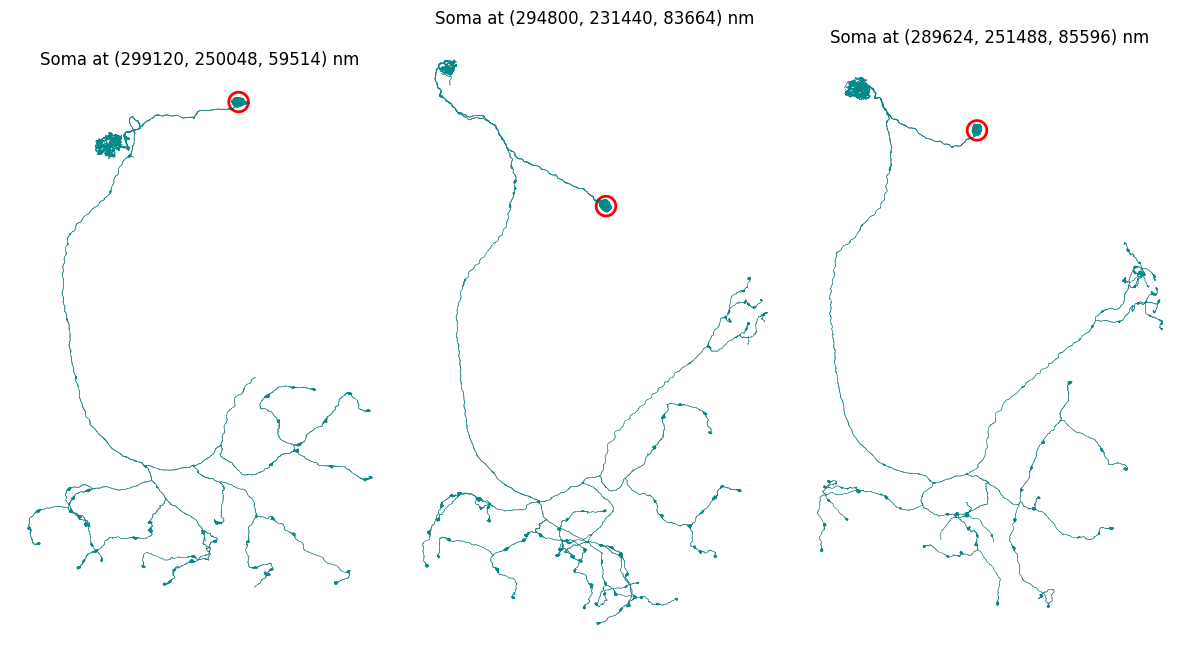
We can also plot the soma location on the mesh, but for this we need to convert from pixels coordinates to nanometers.
from crantpy.utils.config import SCALE_X, SCALE_Y, SCALE_Z
scaling_factors = np.array([SCALE_X, SCALE_Y, SCALE_Z]) # in microns
fig, ax = plt.subplots(1, 3, figsize=(12, 8))
for i in range(3):
# scale soma coordinates to nanometers
soma_nm = [soma_coords_batch[i][0] * SCALE_X, soma_coords_batch[i][1] * SCALE_Y, soma_coords_batch[i][2] * SCALE_Z]
# get the mesh
mesh = mesh_neurons[i]
# plot the mesh with soma location
navis.plot2d(mesh, view=("x", "y"), color='darkcyan', ax=ax[i])
plt.grid(False)
ax[i].set_axis_off()
ax[i].scatter(soma_nm[0], soma_nm[1], color='red', s=200, marker='o', linewidth=2, facecolors='none')
ax[i].set_title(f"Soma at ({int(soma_nm[0])}, {int(soma_nm[1])}, {int(soma_nm[2])}) nm", fontsize=12)
plt.tight_layout()
plt.show()
4. Skeleton Representations#
Skeletons are tree-like representations that capture the branching structure of neurons while being much more computationally efficient than meshes.
Precomputed Skeletons#
CRANTpy provides access to precomputed skeletons stored in the database.
# Fetch precomputed skeletons
skeletons = cp.get_skeletons(sample_ids, progress=True, omit_failures=True)
print(f"Fetched {len(skeletons)} skeletons")
# Examine first skeleton
if len(skeletons) > 0:
skel = skeletons[0]
print(f"\nSkeleton {skel.id} properties:")
print(f" Nodes: {skel.n_nodes:,}")
print(f" Cable length: {skel.cable_length:.2f} nm")
print(f" Branch points: {skel.n_branch_points}")
print(f" End points: {skel.n_endpoints}")
Fetched 3 skeletons
Skeleton 576460752700282748 properties:
Nodes: 221
Cable length: 1075895.12 nm
Branch points: 0
End points: None
L2 Graph-Based Skeletons#
For higher quality morphology, CRANTpy can generate skeletons from L2 chunks. These are often more accurate than precomputed skeletons.
# Get L2 skeleton for a single neuron
l2_skel = cp.get_l2_skeleton(sample_ids[0], refine=True, progress=True)
print(f"L2 skeleton properties:")
print(f" Nodes: {l2_skel.n_nodes:,}")
print(f" Cable length: {l2_skel.cable_length:.2f} nm")
print(f" Branch points: {l2_skel.n_branch_points}")
print(f" End points: {l2_skel.n_endpoints}")
L2 skeleton properties:
Nodes: 309
Cable length: 1409182.00 nm
Branch points: 55
End points: None
# Get L2 skeletons for multiple neurons
l2_skels = cp.get_l2_skeleton(
sample_ids,
refine=True,
progress=True,
max_threads=3,
omit_failures=True
)
print(f"\nFetched {len(l2_skels)} L2 skeletons")
# Compare with precomputed skeletons
if len(skeletons) > 0 and len(l2_skels) > 0:
print(f"\nComparison (first neuron):")
print(f" Precomputed: {skeletons[0].cable_length:.2f} nm")
print(f" L2-based: {l2_skels[0].cable_length:.2f} nm")
Fetched 3 L2 skeletons
Comparison (first neuron):
Precomputed: 1075895.12 nm
L2-based: 1409182.00 nm
L2 Information and Statistics#
You can query L2 chunk information directly for fast morphology estimates. Note that the length_um is an underestimated sum of max diameters across L2 chunks, not a full cable length.
# Get L2 info (fast estimate of morphology)
l2_info = cp.get_l2_info(sample_ids[:5], progress=True)
print("L2 chunk information:")
display(l2_info[['root_id', 'l2_chunks', 'length_um', 'chunks_missing']])
L2 chunk information:
| root_id | l2_chunks | length_um | chunks_missing | |
|---|---|---|---|---|
| 0 | 576460752700282748 | 309 | 39.526 | 0 |
| 1 | 576460752722405178 | 377 | 45.559 | 0 |
| 2 | 576460752681552812 | 390 | 53.886 | 0 |
# Get detailed L2 chunk info
l2_chunk_info = cp.get_l2_chunk_info(sample_ids[0], progress=True)
print(f"\nDetailed L2 chunk info for neuron {sample_ids[0]}:")
print(f" Total chunks: {len(l2_chunk_info)}")
print(f" Columns: {list(l2_chunk_info.columns)}")
display(l2_chunk_info.head())
Detailed L2 chunk info for neuron 576460752700282748:
Total chunks: 309
Columns: ['root_id', 'l2_id', 'x', 'y', 'z', 'vec_x', 'vec_y', 'vec_z', 'size_nm3']
| root_id | l2_id | x | y | z | vec_x | vec_y | vec_z | size_nm3 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 576460752700282748 | 145804519643567814 | 27616 | 9700 | 2815 | 0.404541 | 0.913086 | 0.054138 | 138808320 |
| 1 | 576460752700282748 | 145804519710680256 | 27586 | 9700 | 2816 | -0.446289 | 0.60791 | 0.656738 | 483076608 |
| 2 | 576460752700282748 | 145804588430144816 | 27138 | 10418 | 3038 | -0.220215 | 0.436035 | 0.872559 | 1065555456 |
| 3 | 576460752700282748 | 145804588497240618 | 27602 | 11262 | 3151 | 0.640625 | 0.618652 | 0.455078 | 568780800 |
| 4 | 576460752700282748 | 145804657216724259 | 27596 | 11264 | 3151 | 0.862305 | 0.304199 | 0.405029 | 333666816 |
Finding Anchor Locations#
Find representative coordinates (in pixels) for neurons (useful for annotations or quick localization).
# Find anchor locations
anchor_locs = cp.find_anchor_loc(sample_ids, progress=True)
print("Anchor locations:")
display(anchor_locs)
Anchor locations:
| root_id | x | y | z | |
|---|---|---|---|---|
| 0 | 576460752700282748 | 37256 | 31210 | 1415 |
| 1 | 576460752681552812 | 36862 | 29080 | 1987 |
| 2 | 576460752722405178 | 36214 | 31498 | 2047 |
Rerooting Skeletons to the Soma#
By default, skeletons may not be rooted at the soma. We can reroot them using the detected soma location with the reroot_at_soma() utility function.
Utility Functions for Spatial Operations#
CRANTpy provides convenient utility functions for common spatial operations on skeletons.
# 1. map_position_to_node - Find the nearest skeleton node to any position
position_nm = [240000, 85000, 96000] # Position in nanometers
node_id = cp.map_position_to_node(l2_skels[0], position_nm)
print(f"Position {position_nm} maps to node {node_id}")
# Can also get the distance
node_id, distance = cp.map_position_to_node(l2_skels[0], position_nm, return_distance=True)
print(f"Distance to nearest node: {distance:.2f} nm ({distance/1000:.2f} μm)")
# 2. reroot_at_soma - Automatically detect soma and reroot
# (We already used this above, but here's how to use it without pre-detected soma)
skel_copy = l2_skels[0].copy()
skel_rerooted = cp.reroot_at_soma(skel_copy, inplace=True)
print(f"\nSkeleton rerooted at node {skel_rerooted.root[0]}")
# 3. Practical example: Map synapse positions to skeleton nodes
# This is useful for synapse-to-skeleton mapping (as used in attach_synapses)
synapse_position = [242896, 85536, 96306] # Example synapse coordinate in nm
synapse_node = cp.map_position_to_node(l2_skels[0], synapse_position)
print(f"\nSynapse at {synapse_position} would map to node {synapse_node}")
Position [240000, 85000, 96000] maps to node 11
Distance to nearest node: 6027.00 nm (6.03 μm)
Skeleton rerooted at node 223
Synapse at [242896, 85536, 96306] would map to node 11
# Reroot skeletons at soma location using the convenience function
# This automatically detects soma and reroots at the nearest node
l2_skels_rerooted = cp.reroot_at_soma(l2_skels[:3], progress=True, inplace=False)
print("\nRerooting results:")
for original, rerooted in zip(l2_skels[:3], l2_skels_rerooted):
print(f"Neuron {original.id}: root changed from {original.root[0]} to {rerooted.root[0]}")
Detecting soma: 0%| | 0/3 [00:00<?, ?it/s]
Detecting soma: 33%|███▎ | 1/3 [00:05<00:11, 5.61s/it]
Detecting soma: 67%|██████▋ | 2/3 [00:13<00:07, 7.06s/it]
Rerooting results:
Neuron 576460752700282748: root changed from 0 to 223
Neuron 576460752681552812: root changed from 0 to 276
Neuron 576460752722405178: root changed from 0 to 239
5. Dotprops - Point Cloud Representations#
Dotprops are lightweight point cloud representations with tangent vectors, ideal for NBLAST comparisons.
# Generate dotprops from L2 chunks
dotprops = cp.get_l2_dotprops(
sample_ids,
progress=True,
omit_failures=True
)
print(f"Generated {len(dotprops)} dotprops")
if len(dotprops) > 0:
dp = dotprops[0]
print(f"\nDotprop {dp.id} properties:")
print(f" Points: {len(dp.points)}")
print(f" Has vectors: {dp.vect is not None}")
Generated 3 dotprops
Dotprop 576460752700282748 properties:
Points: 241
Has vectors: True
6. Morphometric Analysis#
Navis provides extensive morphometric analysis capabilities. Let’s explore key metrics.
Basic Morphometrics#
# Use L2 skeletons for morphometric analysis
if len(l2_skels_rerooted) > 0:
morphometrics = []
for skel in l2_skels_rerooted:
metrics = {
'root_id': skel.id,
'cable_length_nm': skel.cable_length,
'n_nodes': skel.n_nodes,
'n_branch_points': skel.n_branch_points,
'n_endpoints': skel.n_endpoints,
'n_branches': len(skel.small_segments),
}
morphometrics.append(metrics)
morph_df = pd.DataFrame(morphometrics)
print("Basic morphometrics:")
display(morph_df)
Basic morphometrics:
| root_id | cable_length_nm | n_nodes | n_branch_points | n_endpoints | n_branches | |
|---|---|---|---|---|---|---|
| 0 | 576460752700282748 | 1409182.000 | 309 | 55 | None | 152 |
| 1 | 576460752681552812 | 1831399.000 | 390 | 68 | None | 183 |
| 2 | 576460752722405178 | 1539621.375 | 377 | 70 | None | 203 |
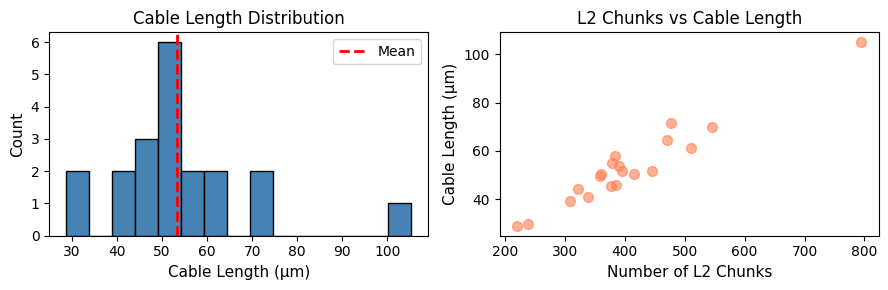
Population-Level Morphological Analysis#
Let’s analyze morphology across a population of neurons.
# Get a larger sample of neurons
opn_sample = opn_ids[:20]
# Get L2 info (fast morphology estimate)
population_l2_info = cp.get_l2_info(opn_sample, progress=True)
print(f"Population morphology statistics:")
print(f"\nCable length (μm):")
print(f" Mean: {population_l2_info['length_um'].mean():.2f}")
print(f" Std: {population_l2_info['length_um'].std():.2f}")
print(f" Range: {population_l2_info['length_um'].min():.2f} - {population_l2_info['length_um'].max():.2f}")
print(f"\nL2 chunks:")
print(f" Mean: {population_l2_info['l2_chunks'].mean():.0f}")
print(f" Range: {population_l2_info['l2_chunks'].min():.0f} - {population_l2_info['l2_chunks'].max():.0f}")
Population morphology statistics:
Cable length (μm):
Mean: 53.39
Std: 16.61
Range: 28.83 - 105.19
L2 chunks:
Mean: 406
Range: 220 - 795
# Visualize population morphology distribution
fig, axes = plt.subplots(1, 2, figsize=(9, 3))
# Cable length distribution
axes[0].hist(population_l2_info['length_um'], bins=15, color='steelblue', edgecolor='black')
axes[0].set_xlabel('Cable Length (μm)', fontsize=11)
axes[0].set_ylabel('Count', fontsize=11)
axes[0].set_title('Cable Length Distribution', fontsize=12)
axes[0].axvline(population_l2_info['length_um'].mean(), color='red', linestyle='--', linewidth=2, label='Mean')
axes[0].legend()
# L2 chunks vs cable length
axes[1].scatter(population_l2_info['l2_chunks'], population_l2_info['length_um'],
color='coral', s=50, alpha=0.6)
axes[1].set_xlabel('Number of L2 Chunks', fontsize=11)
axes[1].set_ylabel('Cable Length (μm)', fontsize=11)
axes[1].set_title('L2 Chunks vs Cable Length', fontsize=12)
plt.tight_layout()
plt.show()
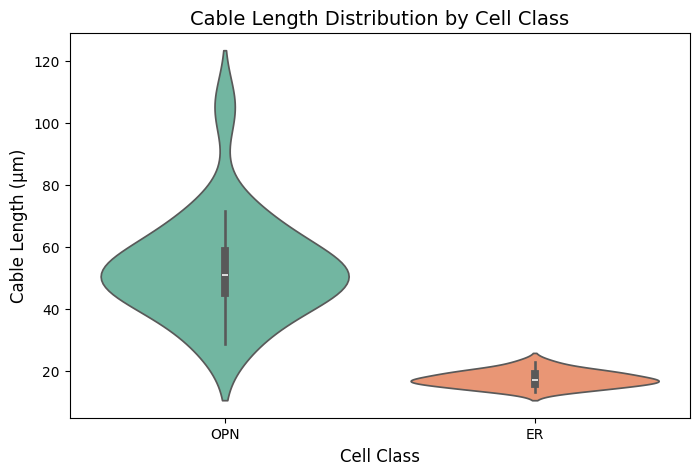
Comparing Cell Types#
# Compare morphology across different cell classes
er_neurons = cp.NeuronCriteria(cell_class='ER')
er_ids = er_neurons.get_roots()[:20]
# Get L2 info for both populations
opn_morph = cp.get_l2_info(opn_sample, progress=True)
opn_morph['cell_class'] = 'OPN'
er_morph = cp.get_l2_info(er_ids, progress=True)
er_morph['cell_class'] = 'ER'
# Combine
combined_morph = pd.concat([opn_morph, er_morph], ignore_index=True)
print("Cable length statistics by cell class:")
print(combined_morph.groupby('cell_class')['length_um'].describe())
# plot the distribution of cable lengths by cell class
plt.figure(figsize=(8, 5))
sns.violinplot(x='cell_class', y='length_um', data=combined_morph, palette='Set2')
plt.title('Cable Length Distribution by Cell Class', fontsize=14)
plt.xlabel('Cell Class', fontsize=12)
plt.ylabel('Cable Length (μm)', fontsize=12)
plt.grid(False)
plt.show()
Cable length statistics by cell class:
count mean std min 25% 50% 75% \
cell_class
ER 20.0 17.4658 2.524752 13.349 15.96050 16.9815 19.0910
OPN 20.0 53.3934 16.605328 28.827 45.23075 51.0715 58.7805
max
cell_class
ER 23.053
OPN 105.185
8. Distance Calculations#
Understanding distances within neurons is crucial for many analyses. We’ll explore both Euclidean (straight-line) and geodesic (along-the-arbor) distances.
Sampling Rate (Node-to-Parent Distances)#
First, let’s calculate the average distance between nodes and their parents - this tells us about the skeleton’s sampling rate:
skel = l2_skels_rerooted[0]
# Get nodes but remove the root (has no parent)
nodes = skel.nodes[skel.nodes.parent_id >= 0]
# Get the x/y/z coordinates of all nodes (except root)
node_locs = nodes[['x', 'y', 'z']].values
# For each node, get its parent's location
parent_locs = skel.nodes.set_index('node_id').loc[nodes.parent_id.values, ['x', 'y', 'z']].values
# Calculate Euclidean distances
distances = np.sqrt(np.sum((node_locs - parent_locs) ** 2, axis=1))
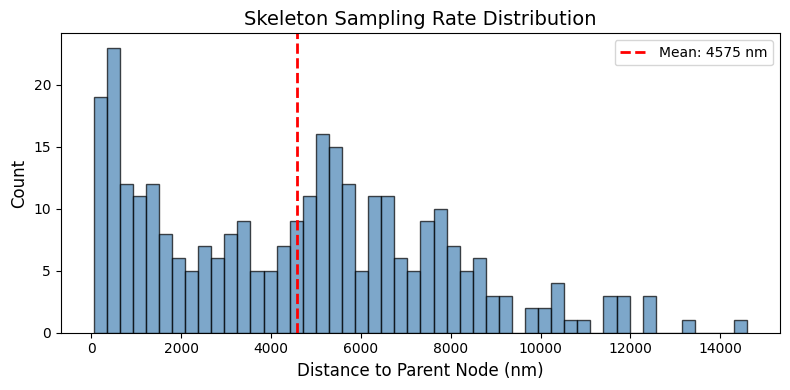
print(f"Sampling rate statistics:")
print(f" Mean distance between nodes: {np.mean(distances):.2f} nm")
print(f" Std: {np.std(distances):.2f} nm")
print(f" Median: {np.median(distances):.2f} nm")
# Plot distribution
plt.figure(figsize=(8, 4))
plt.hist(distances, bins=50, color='steelblue', edgecolor='black', alpha=0.7)
plt.xlabel('Distance to Parent Node (nm)', fontsize=12)
plt.ylabel('Count', fontsize=12)
plt.title('Skeleton Sampling Rate Distribution', fontsize=14)
plt.axvline(np.mean(distances), color='red', linestyle='--', linewidth=2, label=f'Mean: {np.mean(distances):.0f} nm')
plt.legend()
plt.tight_layout()
plt.show()
Sampling rate statistics:
Mean distance between nodes: 4575.27 nm
Std: 3191.86 nm
Median: 4793.04 nm
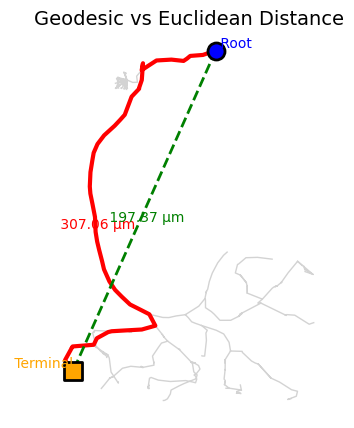
Geodesic Distance: Single Node-to-Node Query#
For distances along the neuron’s branching structure, we need geodesic distances. Let’s calculate the distance from the soma (root) to a terminal node:
# Pick a terminal node
end_nodes = skel.nodes[skel.nodes.type == 'end'].node_id.values
terminal_node = end_nodes[0]
# Calculate geodesic distance from root to terminal node
d_geo = navis.dist_between(skel, skel.root[0], terminal_node)
# Also calculate Euclidean distance for comparison
end_loc = skel.nodes.set_index('node_id').loc[terminal_node, ['x', 'y', 'z']].values
root_loc = skel.nodes.set_index('node_id').loc[skel.root[0], ['x', 'y', 'z']].values
d_eucl = np.sqrt(np.sum((end_loc - root_loc) ** 2))
print(f"Distance from root to terminal node {terminal_node}:")
print(f" Geodesic (along arbor): {d_geo:.2f} nm ({d_geo/1000:.2f} μm)")
print(f" Euclidean (straight line): {d_eucl:.2f} nm ({d_eucl/1000:.2f} μm)")
print(f" Ratio (tortuosity): {d_geo/d_eucl:.2f}x")
Distance from root to terminal node 5:
Geodesic (along arbor): 307062.21 nm (307.06 μm)
Euclidean (straight line): 197367.96 nm (197.37 μm)
Ratio (tortuosity): 1.56x
# Visualize the difference between geodesic and Euclidean distances
# Find the shortest path along the skeleton
path = nx.shortest_path(skel.graph.to_undirected(), skel.root[0], terminal_node)
# Get coordinates for the path
path_co = skel.nodes.set_index('node_id').loc[path, ['x', 'y', 'z']].copy()
# Plot the neuron
fig, ax = navis.plot2d(skel, color='lightgray', method='2d', view=('x', 'y'), figsize=(5, 5))
# Add geodesic path (along the skeleton)
ax.plot(path_co.x, path_co.y, color='red', linewidth=3, label='Geodesic path', zorder=10)
# Add Euclidean path (straight line)
ax.plot([root_loc[0], end_loc[0]], [root_loc[1], end_loc[1]],
color='green', linewidth=2, linestyle='--', label='Euclidean path', zorder=10)
# Mark root and terminal node
ax.scatter(root_loc[0], root_loc[1], s=150, color='blue', marker='o',
label='Root (soma)', zorder=15, edgecolors='black', linewidths=2)
ax.scatter(end_loc[0], end_loc[1], s=150, color='orange', marker='s',
label='Terminal node', zorder=15, edgecolors='black', linewidths=2)
# Add text annotations
ax.text(root_loc[0], root_loc[1], ' Root', color='blue', fontsize=10, verticalalignment='bottom', horizontalalignment='left', zorder=20)
ax.text(end_loc[0], end_loc[1], ' Terminal', color='orange', fontsize=10, verticalalignment='bottom', horizontalalignment='right', zorder=20)
# add distance annotations
mid_geo = path_co.iloc[len(path_co)//2]
mid_eucl = [(root_loc[0] + end_loc[0]) / 2, (root_loc[1] + end_loc[1]) / 2]
ax.text(mid_geo.x, mid_geo.y, f' {d_geo/1000:.2f} μm', color='red', fontsize=10, verticalalignment='bottom', horizontalalignment='center', zorder=20)
ax.text(mid_eucl[0], mid_eucl[1], f' {d_eucl/1000:.2f} μm', color='green', fontsize=10, verticalalignment='top', horizontalalignment='center', zorder=20)
ax.set_title('Geodesic vs Euclidean Distance', fontsize=14)
ax.set_axis_off()
plt.show()
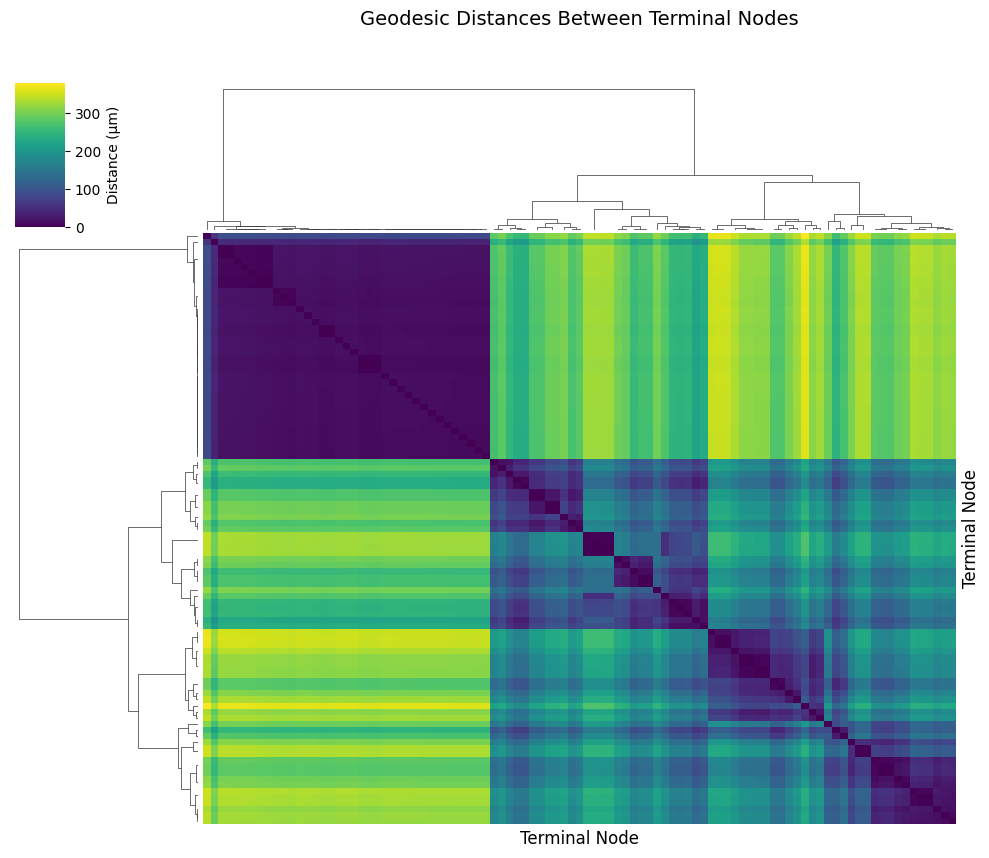
Geodesic Distance Matrix#
For analyzing distances across many nodes, we can use navis.geodesic_matrix() to compute a distance matrix. Let’s calculate distances between all terminal nodes:
# Calculate distances from all end nodes to all other nodes
ends = skel.nodes[skel.nodes.type == 'end'].node_id.values
print(f"Calculating geodesic distances for {len(ends)} terminal nodes...")
# Get the distance matrix
dist_matrix = navis.geodesic_matrix(skel, from_=ends)
# Subset to only end-to-end distances
dist_matrix_ends = dist_matrix.loc[ends, ends]
print(f"\nDistance matrix shape: {dist_matrix_ends.shape}")
print(f"Distance statistics (μm):")
print(f" Mean: {dist_matrix_ends.values.mean()/1000:.2f}")
print(f" Median: {np.median(dist_matrix_ends.values)/1000:.2f}")
print(f" Max: {dist_matrix_ends.values.max()/1000:.2f}")
display(dist_matrix_ends.head())
Calculating geodesic distances for 97 terminal nodes...
Distance matrix shape: (97, 97)
Distance statistics (μm):
Mean: 194.81
Median: 219.03
Max: 379.44
| 5 | 6 | 7 | 8 | 44 | 12 | 19 | 10 | 39 | 42 | ... | 286 | 282 | 281 | 289 | 290 | 304 | 301 | 307 | 305 | 306 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 0.000000 | 1470.138794 | 21770.945312 | 21924.505859 | 43284.164062 | 82830.328125 | 68335.960938 | 162903.531250 | 155059.750000 | 97709.773438 | ... | 196621.8750 | 194101.625000 | 194177.703125 | 213741.078125 | 215143.28125 | 214334.812500 | 206430.062500 | 228993.875000 | 230859.656250 | 229488.406250 |
| 6 | 1470.138794 | 0.000000 | 21653.496094 | 21807.058594 | 43166.718750 | 82712.882812 | 68218.515625 | 162786.093750 | 154942.312500 | 97592.328125 | ... | 196504.4375 | 193984.187500 | 194060.265625 | 213623.640625 | 215025.84375 | 214217.375000 | 206312.625000 | 228876.437500 | 230742.218750 | 229370.968750 |
| 7 | 21770.945312 | 21653.496094 | 0.000000 | 963.262634 | 22322.921875 | 61869.085938 | 47374.718750 | 141942.296875 | 134098.500000 | 76748.531250 | ... | 175660.6250 | 173140.375000 | 173216.468750 | 192779.843750 | 194182.03125 | 193373.578125 | 185468.812500 | 208032.625000 | 209898.406250 | 208527.156250 |
| 8 | 21924.505859 | 21807.058594 | 963.262634 | 0.000000 | 22476.482422 | 62022.648438 | 47528.281250 | 142095.859375 | 134252.062500 | 76902.093750 | ... | 175814.1875 | 173293.937500 | 173370.031250 | 192933.406250 | 194335.59375 | 193527.140625 | 185622.375000 | 208186.187500 | 210051.968750 | 208680.718750 |
| 44 | 43284.164062 | 43166.718750 | 22322.921875 | 22476.482422 | 0.000000 | 58810.382812 | 44316.015625 | 138883.593750 | 131039.796875 | 73689.828125 | ... | 172601.9375 | 170081.671875 | 170157.750000 | 189721.125000 | 191123.34375 | 190314.875000 | 182410.109375 | 204973.921875 | 206839.703125 | 205468.453125 |
5 rows × 97 columns
# Visualize the distance matrix with hierarchical clustering
from scipy.cluster.hierarchy import linkage
from scipy.spatial.distance import squareform
# Generate linkage from the distances
Z = linkage(squareform(dist_matrix_ends.values/1000, checks=False), method='ward')
# Create clustermap
fig = plt.figure(figsize=(10, 8))
cm = sns.clustermap(
dist_matrix_ends.values/1000, # Convert to microns
cmap='viridis',
col_linkage=Z,
row_linkage=Z,
cbar_kws={'label': 'Distance (μm)'},
figsize=(10, 8)
)
cm.ax_heatmap.set_xlabel('Terminal Node', fontsize=12)
cm.ax_heatmap.set_ylabel('Terminal Node', fontsize=12)
cm.ax_heatmap.set_title('Geodesic Distances Between Terminal Nodes', fontsize=14, pad=150)
# Remove tick labels for cleaner look
cm.ax_heatmap.set_xticks([])
cm.ax_heatmap.set_yticks([])
plt.show()
print("\nNote: Clustering in the heatmap can reveal dendritic vs axonal compartments!")
print("Nodes that cluster together are likely in the same compartment.")
<Figure size 1000x800 with 0 Axes>
Note: Clustering in the heatmap can reveal dendritic vs axonal compartments!
Nodes that cluster together are likely in the same compartment.
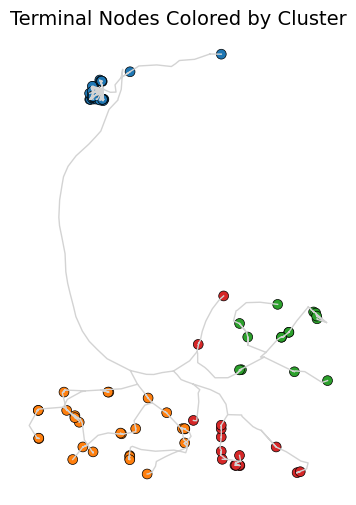
# get the clusters and plot them on the neuron
from scipy.cluster.hierarchy import fcluster
# Cut the dendrogram to form 2 clusters
clusters = fcluster(Z, t=4, criterion='maxclust')
# plot all the nodes colored by cluster
fig, ax = navis.plot2d(skel, method='2d', view=('x', 'y'), color='lightgray', figsize=(6, 6))
node_colors = sns.color_palette('tab10', n_colors=max(clusters)).as_hex()
node_colors = [node_colors[c-1] for c in clusters]
ax.scatter(
skel.nodes.set_index('node_id').loc[ends, 'x'],
skel.nodes.set_index('node_id').loc[ends, 'y'],
c=node_colors, s=50, edgecolors='black', linewidths=0.5, label='Terminal Nodes'
)
ax.set_title('Terminal Nodes Colored by Cluster', fontsize=14)
ax.set_axis_off()
plt.show()
Distance-based Node Coloring#
We can also use distances to color the neuron, highlighting how far each node is from the root:
# Calculate distance from root to all nodes
root_node = skel.root[0]
all_node_ids = skel.nodes.node_id.values
distances_all = navis.geodesic_matrix(skel, from_=all_node_ids)
# Add to nodes dataframe
skel.nodes['root_dist'] = distances_all.loc[root_node, :].values
# set nan to 0
skel.nodes['root_dist'] = skel.nodes['root_dist'].fillna(0)
print(f"Distance from root statistics:")
print(f" Mean: {skel.nodes['root_dist'].mean()/1000:.2f} μm")
print(f" Max: {skel.nodes['root_dist'].max()/1000:.2f} μm")
print(f" Std: {skel.nodes['root_dist'].std()/1000:.2f} μm")
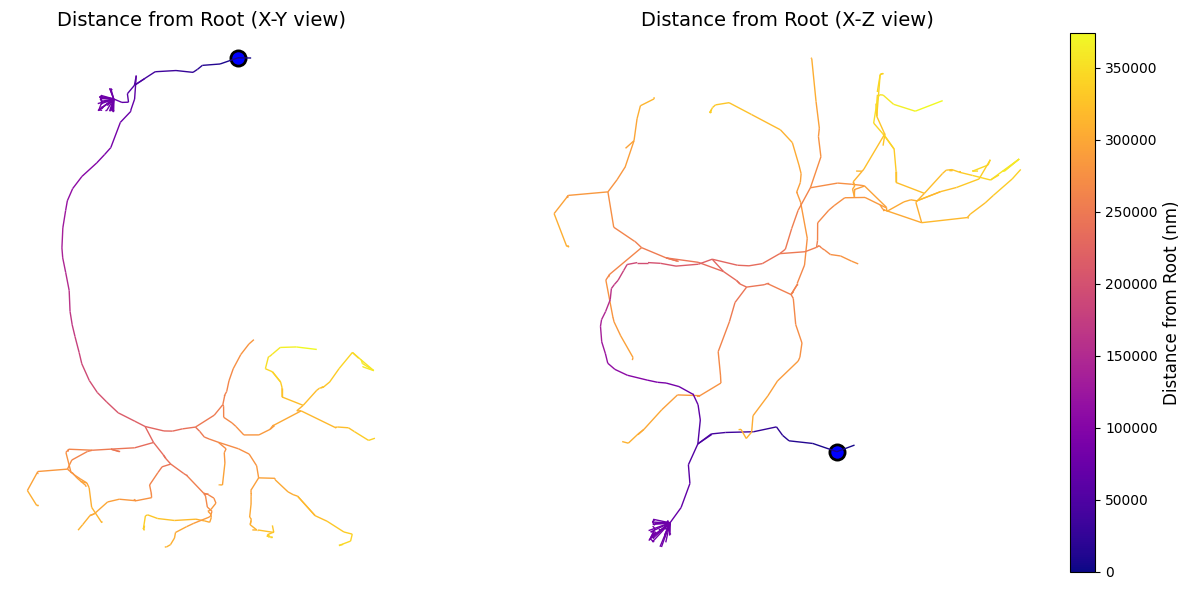
# Plot neuron colored by distance from root
fig, axes = plt.subplots(1, 2, figsize=(14, 6))
# View 1: X-Y
navis.plot2d(
skel,
color_by='root_dist',
palette='plasma',
method='2d',
view=('x', 'y'),
ax=axes[0]
)
# plot the root
axes[0].scatter(
root_loc[0], root_loc[1],
color='blue', s=120, edgecolors='black', linewidths=2, label='Root'
)
axes[0].set_title('Distance from Root (X-Y view)', fontsize=14)
axes[0].set_axis_off()
# View 2: X-Z
navis.plot2d(
skel,
color_by='root_dist',
palette='plasma',
method='2d',
view=('x', 'z'),
ax=axes[1]
)
# plot the root
axes[1].scatter(
root_loc[0], root_loc[2],
color='blue', s=120, edgecolors='black', linewidths=2, label='Root'
)
axes[1].set_title('Distance from Root (X-Z view)', fontsize=14)
axes[1].set_axis_off()
# add colorbar
minimum = skel.nodes['root_dist'].min()
maximum = skel.nodes['root_dist'].max()
cbar = plt.colorbar(plt.cm.ScalarMappable(cmap='plasma', norm=plt.Normalize(vmin=minimum, vmax=maximum)), orientation='vertical', fraction=0.05, pad=0.1, cax=axes[1].inset_axes([1.05, 0, 0.05, 1]))
cbar.set_label('Distance from Root (nm)', fontsize=12)
plt.tight_layout()
plt.show()
Distance from root statistics:
Mean: 236.91 μm
Max: 374.03 μm
Std: 101.03 μm
9. Advanced Morphometric Analysis#
Segment Analysis#
Navis provides segment_analysis() which collects comprehensive morphometrics for each linear segment (between branch points), including Strahler index, cable length, distance to root, and tortuosity.
# Perform comprehensive segment analysis
seg_analysis = navis.segment_analysis(l2_skels_rerooted[0])
print(f"Segment analysis results:")
print(f" Total segments: {len(seg_analysis)}")
print(f"\nFirst few segments:")
display(seg_analysis.head())
print(f"\nSummary statistics:")
display(seg_analysis[['length', 'tortuosity', 'root_dist', 'strahler_index']].describe())
Segment analysis results:
Total segments: 152
First few segments:
| length | tortuosity | root_dist | strahler_index | radius_mean | radius_min | radius_max | volume | |
|---|---|---|---|---|---|---|---|---|
| 0 | 793.792721 | 1.009957 | 306268.419732 | 1 | 221.333333 | 193 | 244 | 1.326052e+08 |
| 1 | 20572.301259 | 1.093203 | 285696.118473 | 2 | 271.000000 | 193 | 314 | 5.024232e+09 |
| 2 | 676.346065 | 1.000000 | 306268.419732 | 1 | 191.000000 | 189 | 193 | 7.751780e+07 |
| 3 | 12285.961101 | 1.034210 | 273410.157373 | 2 | 212.750000 | 117 | 314 | 9.929913e+08 |
| 4 | 404.850590 | 1.000000 | 285696.118473 | 1 | 243.000000 | 172 | 314 | 7.724016e+07 |
Summary statistics:
| length | tortuosity | root_dist | strahler_index | |
|---|---|---|---|---|
| count | 152.000000 | 152.000000 | 152.000000 | 152.000000 |
| mean | 9270.934315 | 1.037665 | 225768.867012 | 1.513158 |
| std | 16940.222298 | 0.100488 | 106298.128510 | 0.813681 |
| min | 55.172457 | 1.000000 | 0.000000 | 1.000000 |
| 25% | 1192.933156 | 1.000000 | 80607.110184 | 1.000000 |
| 50% | 4983.385660 | 1.000000 | 265986.027429 | 1.000000 |
| 75% | 10191.787491 | 1.024821 | 306346.790386 | 2.000000 |
| max | 173550.345434 | 1.788708 | 360682.099962 | 5.000000 |
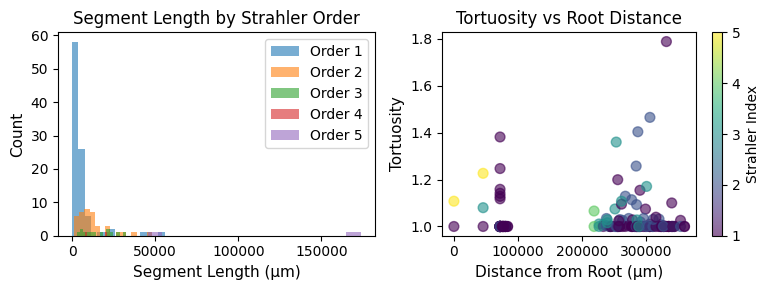
# Visualize segment properties
seg_analysis = navis.segment_analysis(l2_skels_rerooted[0])
fig, axes = plt.subplots(1, 2, figsize=(8, 3))
# Segment length distribution by Strahler order
for order in sorted(seg_analysis['strahler_index'].unique()):
data = seg_analysis[seg_analysis['strahler_index'] == order]['length']
axes[0].hist(data, bins=15, alpha=0.6, label=f'Order {int(order)}')
axes[0].set_xlabel('Segment Length (μm)', fontsize=11)
axes[0].set_ylabel('Count', fontsize=11)
axes[0].set_title('Segment Length by Strahler Order', fontsize=12)
axes[0].legend()
# Tortuosity vs distance from root
scatter = axes[1].scatter(
seg_analysis['root_dist'],
seg_analysis['tortuosity'],
c=seg_analysis['strahler_index'],
cmap='viridis',
s=50,
alpha=0.6
)
axes[1].set_xlabel('Distance from Root (μm)', fontsize=11)
axes[1].set_ylabel('Tortuosity', fontsize=11)
axes[1].set_title('Tortuosity vs Root Distance', fontsize=12)
plt.colorbar(scatter, ax=axes[1], label='Strahler Index')
plt.tight_layout()
plt.show()
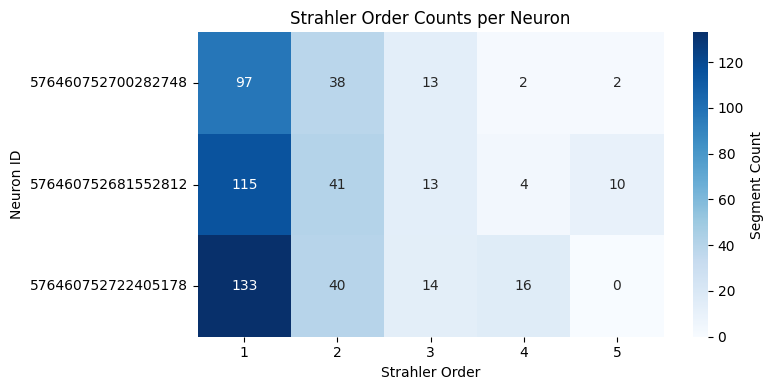
# compare strahler order counts between neurons
# calculate strahler order counts for each neuron
strahler_counts = []
for skel in l2_skels_rerooted:
seg_analysis = navis.segment_analysis(skel)
counts = seg_analysis['strahler_index'].value_counts().to_dict()
counts['root_id'] = skel.id
strahler_counts.append(counts)
strahler_df = pd.DataFrame(strahler_counts).fillna(0).set_index('root_id').astype(int)
# show as a heatmap
plt.figure(figsize=(8, 4))
sns.heatmap(strahler_df, annot=True, fmt='d', cmap='Blues', cbar_kws={'label': 'Segment Count'})
plt.title('Strahler Order Counts per Neuron')
plt.xlabel('Strahler Order')
plt.ylabel('Neuron ID')
plt.tight_layout()
plt.show()
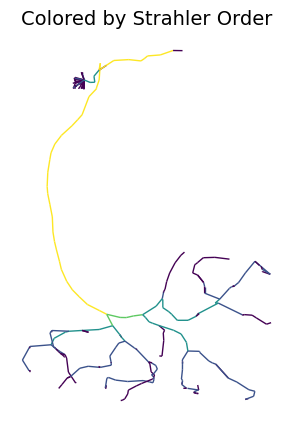
Visualizing Node Properties#
# add strahler index to skeleton nodes
navis.strahler_index(l2_skels_rerooted[0])
# Color by Strahler order
fig, ax = navis.plot2d(
l2_skels_rerooted[0],
view=('x', 'y'),
color_by='strahler_index',
palette='viridis',
method='2d',
figsize=(5, 5)
)
ax.grid(False)
ax.set_title('Colored by Strahler Order', fontsize=14)
ax.set_axis_off()
plt.show()
Sholl analysis#
Sholl analysis quantifies branching complexity as a function of distance from the soma.
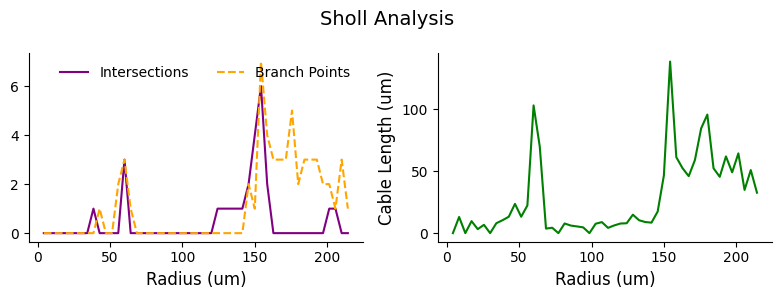
Euclidean distance from soma#
Typically, Sholl analysis is performed using Euclidean distance from the soma.
# perform sholl analysis with euclidean radii of 50 nm increments from the root
sholl_results = navis.sholl_analysis(l2_skels_rerooted[0], radii=50, center='root')
display(sholl_results.head())
# plot sholl results
fig, ax = plt.subplots(1,2, figsize=(8, 3))
ax[0].plot(sholl_results.index/1000, sholl_results['intersections'].values, ls ='-', color='purple', label='Intersections')
ax[0].plot(sholl_results.index/1000, sholl_results['branch_points'].values, ls='--', color='orange', label='Branch Points')
ax[0].set_xlabel('Radius (um)', fontsize=12)
ax[0].spines[['top', 'right']].set_visible(False)
ax[0].legend(frameon=False, ncols = 2)
ax[1].plot(sholl_results.index/1000, sholl_results['cable_length'].values/1000, ls='-', color='green')
ax[1].set_xlabel('Radius (um)', fontsize=12)
ax[1].set_ylabel('Cable Length (um)', fontsize=12)
ax[1].spines[['top', 'right']].set_visible(False)
plt.suptitle('Sholl Analysis', fontsize=14)
plt.tight_layout()
| intersections | cable_length | branch_points | |
|---|---|---|---|
| radius | |||
| 4283.629551 | 0 | 0.000000 | 0 |
| 8567.259101 | 0 | 13065.068359 | 0 |
| 12850.888652 | 0 | 0.000000 | 0 |
| 17134.518202 | 0 | 9701.698242 | 0 |
| 21418.147753 | 0 | 3230.100830 | 0 |
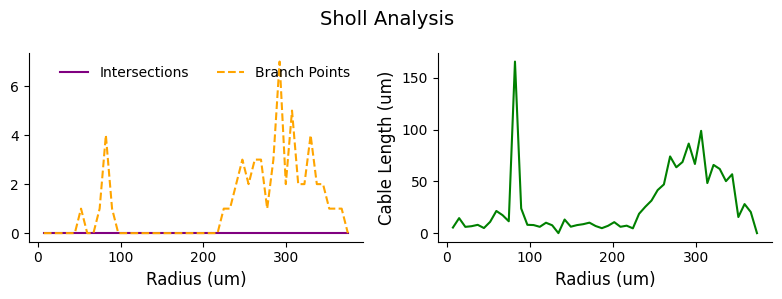
Geodesic distance from soma#
Sholl analysis can also be performed using geodesic distance along the neuron structure.
# perform sholl analysis with euclidean radii of 50 nm increments from the root
sholl_results = navis.sholl_analysis(l2_skels_rerooted[0], radii=50, center='root', geodesic=True)
display(sholl_results.head())
# plot sholl results
fig, ax = plt.subplots(1,2, figsize=(8, 3))
ax[0].plot(sholl_results.index/1000, sholl_results['intersections'].values, ls ='-', color='purple', label='Intersections')
ax[0].plot(sholl_results.index/1000, sholl_results['branch_points'].values, ls='--', color='orange', label='Branch Points')
ax[0].set_xlabel('Radius (um)', fontsize=12)
ax[0].spines[['top', 'right']].set_visible(False)
ax[0].legend(frameon=False, ncols = 2)
ax[1].plot(sholl_results.index/1000, sholl_results['cable_length'].values/1000, ls='-', color='green')
ax[1].set_xlabel('Radius (um)', fontsize=12)
ax[1].set_ylabel('Cable Length (um)', fontsize=12)
ax[1].spines[['top', 'right']].set_visible(False)
plt.suptitle('Sholl Analysis', fontsize=14)
plt.tight_layout()
| intersections | cable_length | branch_points | |
|---|---|---|---|
| radius | |||
| 7480.571289 | 0 | 5413.287598 | 0 |
| 14961.142578 | 0 | 14514.019531 | 0 |
| 22441.714844 | 0 | 6069.560547 | 0 |
| 29922.285156 | 0 | 6710.543457 | 0 |
| 37402.855469 | 0 | 8032.163574 | 0 |
Summary#
In this comprehensive deep dive, you learned:
✅ Mesh representations - Fetching and analyzing neuron surface meshes
✅ Skeleton representations - Multiple methods (precomputed, L2-based)
✅ L2 graph analysis - High-quality morphology from L2 chunks
✅ Dotprops - Lightweight representations for fast comparisons
✅ Morphometric analysis - Cable length, branching, Strahler order, tortuosity
✅ Soma detection - Automated identification from mesh and skeleton data
✅ Visualization - 2D, 3D, and property-based coloring
✅ Population analysis - Comparing morphology across neuron groups
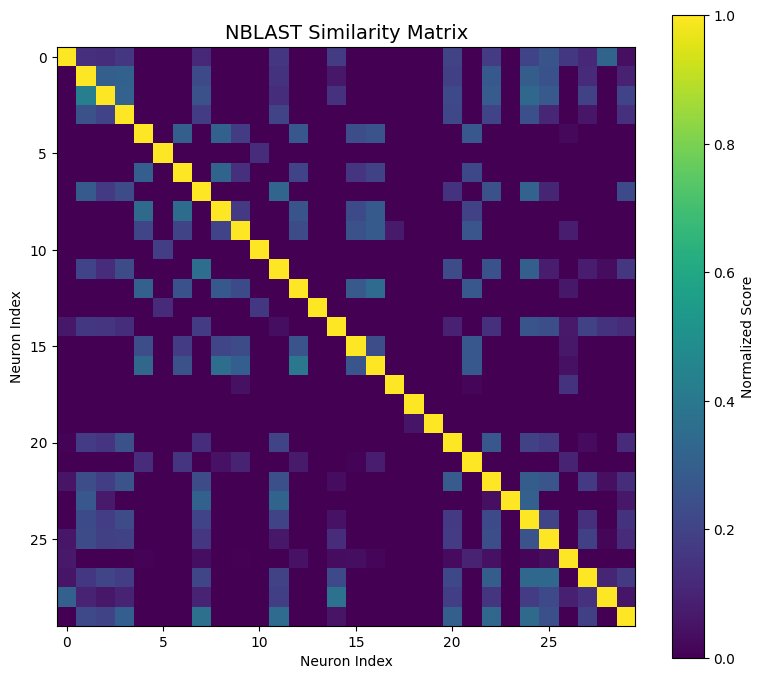
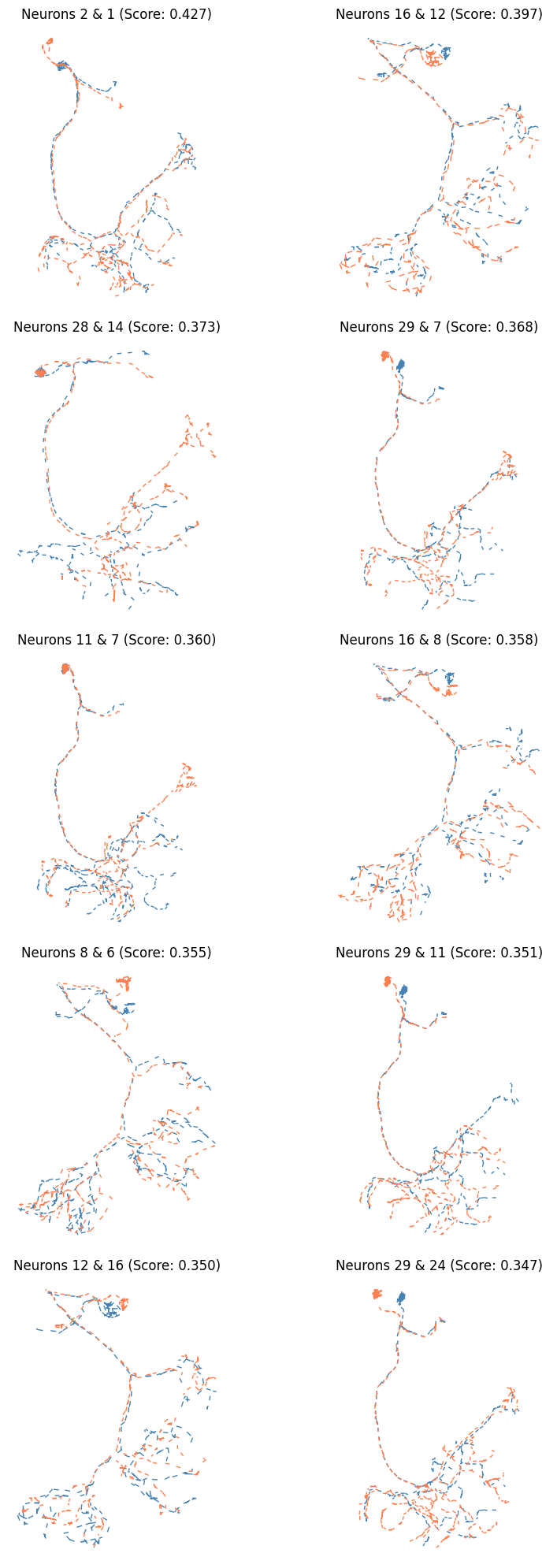
✅ Navis integration - Advanced analysis (NBLAST, resampling, smoothing)
Key Takeaways#
Choose the right representation: Meshes for detail, skeletons for analysis, dotprops for comparison
L2-based morphology: Often more accurate than precomputed data
Fast estimates: Use
get_l2_info()for quick morphology overviewLeverage navis: Full access to navis’s morphometric toolkit
Population studies: Compare morphology across cell types and conditions